Anti-c-Myc: Mouse c-Myc Antibody |
 |
BACKGROUND Members of the Myc/Max/Mad network function as transcriptional regulators with roles in various aspects of cell behavior including proliferation, differentiation and apoptosis (1). These proteins share a common basic-helix-loop-helix leucine zipper (bHLH-ZIP) motif required for dimerization and DNA-binding. Max was originally discovered based on its ability to associate with cMyc and found to be required for the ability of Myc to bind DNA and activate transcription (2). Subsequently, Max has been viewed as a central component of the transcriptional network, forming homodimers as well as heterodimers with other members of the Myc and Mad families (1). The association between Max and either Myc or Mad can have opposing effects on transcriptional regulation and cell behavior (1). The Mad family consists of four related proteins; Mad1, Mad2 (Mxi1), Mad3 and Mad4, and the more distantly related members of the bHLH-ZIP family, Mnt and Mga. Like Myc, the Mad proteins are tightly regulated with short half-lives. In general, Mad family members interfere with Myc-mediated processes such as proliferation, transformation and prevention of apoptosis by inhibiting transcription (3, 4).
REFERENCES
1. Baudino, T.A. and Cleveland, J.L. (2001) Mol. Cell. Biol.21, 691-702.
2. Blackwood, E.M. and Eisenman, R.N. (1991) Science251, 1211-1217.
3. Henriksson, M. and Lüscher, B. (1996) Adv. Cancer Res.68, 109-182.
4. Grandori, C. et al. (2000) Annu. Rev. Cell Dev. Biol.16, 653-699
Products are for research use only. They are not intended for human, animal, or diagnostic applications.
Параметры
| Cat.No.: | CC10042 |
| Antigen: | Recombinant c-Myc expressed in mammalian cells. |
| Isotype: | Mouse IgG |
Species & predicted species cross- reactivity ( ): | Human, Mouse |
Applications & Suggested starting dilutions:* | WB 1:1000 IP n/d IHC 1:100 ICC n/d FACS n/d |
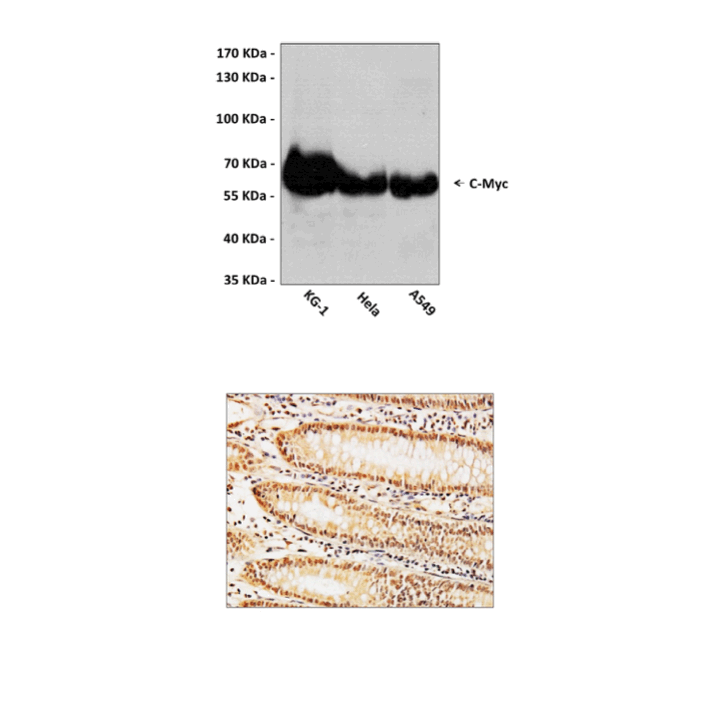
Predicted Molecular Weight of protein: | 57-65 kDa |
| Specificity/Sensitivity: | Detects endogenous c-Myc proteins without cross-reactivity with other family members. |
| Storage: | Store at -20°C, 4°C for frequent use. Avoid repeated freeze-thaw cycles. |
*Optimal working dilutions must be determined by end user.
Информация представлена исключительно в ознакомительных целях и ни при каких условиях не является публичной офертой