Anti-CLOCK: Rabbit CLOCK Antibody |
 |
BACKGROUND Circadian rhythms govern many key physiological processes that fluctuate with a period of approximately 24 hours. These processes include the sleep-wake cycle, glucose, lipid and drug metabolism, heart rate, hormone secretion, renal blood flow, and body temperature, as well as basic cellular processes such as DNA repair and the timing of the cell division cycle (1, 2). The mammalian circadian system consists of many individual tissue-specific clocks (peripheral clocks) that are controlled by a master circadian pacemaker residing in the suprachiasmatic nuclei (SCN) of the brain (1, 2). The periodic circadian rhythm is prominently manifested by the light-dark cycle, which is sensed by the visual system and processed by the SCN. The SCN processes the light-dark information and synchronizes peripheral clocks through neural and humoral output signals (1, 2).The cellular circadian clockwork consists of interwoven positive and negative regulatory loops, or limbs (1, 2). The positive limb includes the CLOCK and BMAL1 proteins, two basic helix-loop-helix-PAS containing transcription factors that bind E box enhancer elements and activate transcription of their target genes. CLOCK is a histone acetyltransferase (HAT) protein, which acetylates both histone H3 and H4 (3). BMAL1 binds to CLOCK and enhances its HAT activity (3). The CLOCK/BMAL1 dimer exhibits a periodic oscillation in both nuclear/cytoplasmic localization and protein levels, both of which are regulated by phosphorylation (4, 5). CLOCK/BMAL1 target genes include the Cry and Per genes, whose proteins form the negative limb of the circadian clockwork system (1, 2). CRY and PER proteins (CRY1, CRY2, PER1, PER2 and PER3) form oligomers that also periodically shuttle between the nucleus and cytoplasm. When in the nucleus, CRY/PER proteins inhibit CLOCK/BMAL1-mediated transcriptional activation, thus completing the circadian transcriptional loop (1, 2). In tissues, roughly six to eight percent of all genes exhibit a circadian expression pattern (1, 2). This 24-hour periodicity in gene expression results from coordination of the positive and negative regulatory limbs of the cellular clockwork system, and is fine-tuned by outside signals received from the SCN.
REFERENCES
1. Albrecht, U. and Eichele, G. (2003) Curr Opin Genet Dev13, 271-7.
2. Virshup, D.M. et al. (2007) Cold Spring Harb Symp Quant Biol72, 413-20.
3. Doi, M. et al. (2006) Cell125, 497-508.
4. Kondratov, R.V. et al. (2003) Genes Dev17, 1921-32.
5. Kwon, I. et al. (2006) Mol Cell Biol26, 7318-30.
Products are for research use only. They are not intended for human, animal, or diagnostic applications.
Параметры
| Cat.No.: | CC1057 |
| Antigen: | Recombinant mouse CLOCK expressed in mammalian cells. |
| Isotype: | Rabbit IgG |
Species & predicted species cross- reactivity ( ): | Human, Mouse |
Applications & Suggested starting dilutions:* | WB 1:100 IP n/d IHC n/d ICC n/d FACS n/d |
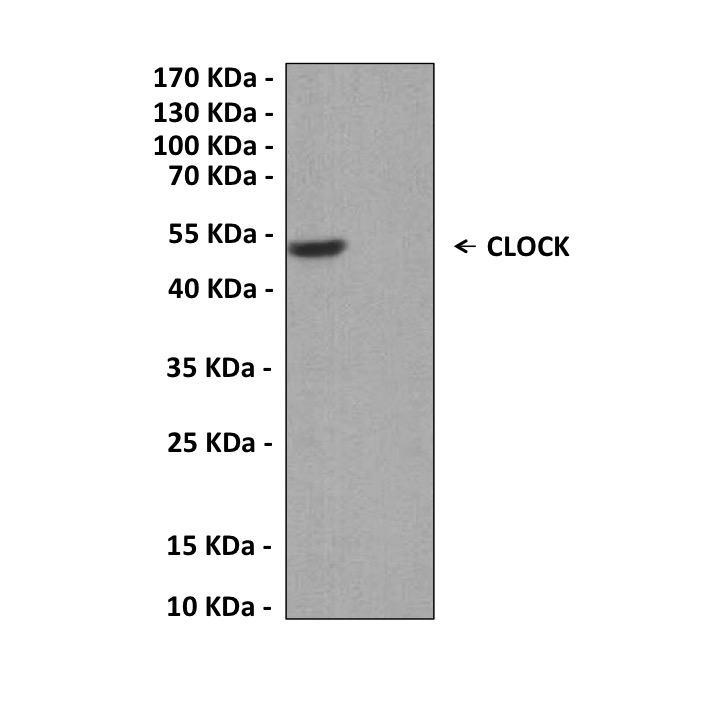
Predicted Molecular Weight of protein: | 100 kDa |
| Specificity/Sensitivity: | Detects endogenous CLOCK proteins without cross-reactivity with other family members. |
| Storage: | Store at -20°C, 4°C for frequent use. Avoid repeated freeze-thaw cycles. |
*Optimal working dilutions must be determined by end user.
Информация представлена исключительно в ознакомительных целях и ни при каких условиях не является публичной офертой