Anti-CTCF: Mouse CTCF Antibody |
 |
BACKGROUND CTCF is a ubiquitous, 11-zinc-finger DNA-binding protein involved in transcriptional regulation, reading of imprinted sites, X chromosome inactivation, and enhancer blocking/insulator function. Numerous studies highlight evidence for CTCF-mediated intra- and interchromosomal contacts at several developmentally regulated genomic loci. It has been demonstrated that CTCF plays primary role in the global organization of chromatin architecture and CTCF may be a heritable component of an epigenetic system regulating the interplay between DNA methylation, higher-order chromatin structure, and lineage-specific gene expression.1 In vertebrates, a role for CTCF in reading and propagating epigenetic marks has been shown.
Eukaryotic genomes are organized into functional units containing individual genes or gene groups together with the corresponding regulatory elements. These functional units have to be insulated from each other in order to prevent illegitimate interactions with other transcriptional units. One particular aspect of insulation is enhancer blocking. In higher eukaryotes, several insulator sequences have been identified. A common model for the mechanism of enhancer blocking predicts association of looped chromatin domains with the nuclear matrix. CTCF has unusual properties, exerting an influence on local chromatin architecture through the formation of higher order structures. It also has the property, when located between a promoter and enhancer, of blocking enhancer function, potentially through its ability to organize chromosomal domains within the nucleus. Furthermore, it was shown that not only CTCF chromatin binding, but also 'boundary type' long-range chromatin interaction can be detected in mitotic chromatin, whereas the enhancer/promoter interaction at the same locus is lost in mitosis.2 Through its 11 zinc fingers CTCF can bind to a range of DNA sequences, to itself, and to nuclear structural proteins such as nucleophosmin and matrix attachment regions (MARs). Genome scans of CTCF enrichment show that CTCF sites are overrepresented in genes and promoter regions, with only 41–56% at intergenic locations. Thus, although CTCF sites are proposed to be insulators, their in vivo functions may not be limited to insulation. In the Igf2/H19 locus, CTCF binding to the imprinting control region on the maternal allele is required to loop out the Igf2 gene and prevent interaction between Igf2 and enhancers. Similarly, CTCF binding and chromatin organization is necessary for activation of IFNG during T-cell differentiation. However, in the mouse β-globin locus, CTCF binding at the flanking HS5 and 3′HS1 sites results in loop formation, but the loop does not appear to be required for normal globin gene regulation in erythroid progenitor cells. These functions of CTCF all involve CTCF-mediated long-range chromatin organization, which can be manifest intra- or interchromosomally. CTCF is a very strong candidate for the role of coordinating the expression level of coding sequences with their three-dimensional position in the nucleus, apparently responding to a “code” in the DNA itself. Dynamic interactions between chromatin fibers in the context of nuclear architecture have been implicated in various aspects of genome functions. CTCF-chromatin interaction mediates gene regulation in the three-dimensional nuclear space. The “CTCF code,” explaining the mechanistic basis of how the information encrypted in DNA may be interpreted by CTCF into diverse nuclear functions.3 Furthermore, CTCF binds DNA in a methylation-sensitive manner. Therefore, DNA methylation has the potential to positively regulate gene transcription, albeit in an indirect manner, by preventing CTCF binding and thereby abolishing an enhancer block.4 In cancer, CTCF appears to function as a tumor suppressor gene.
Eukaryotic genomes are organized into functional units containing individual genes or gene groups together with the corresponding regulatory elements. These functional units have to be insulated from each other in order to prevent illegitimate interactions with other transcriptional units. One particular aspect of insulation is enhancer blocking. In higher eukaryotes, several insulator sequences have been identified. A common model for the mechanism of enhancer blocking predicts association of looped chromatin domains with the nuclear matrix. CTCF has unusual properties, exerting an influence on local chromatin architecture through the formation of higher order structures. It also has the property, when located between a promoter and enhancer, of blocking enhancer function, potentially through its ability to organize chromosomal domains within the nucleus. Furthermore, it was shown that not only CTCF chromatin binding, but also 'boundary type' long-range chromatin interaction can be detected in mitotic chromatin, whereas the enhancer/promoter interaction at the same locus is lost in mitosis.2 Through its 11 zinc fingers CTCF can bind to a range of DNA sequences, to itself, and to nuclear structural proteins such as nucleophosmin and matrix attachment regions (MARs). Genome scans of CTCF enrichment show that CTCF sites are overrepresented in genes and promoter regions, with only 41–56% at intergenic locations. Thus, although CTCF sites are proposed to be insulators, their in vivo functions may not be limited to insulation. In the Igf2/H19 locus, CTCF binding to the imprinting control region on the maternal allele is required to loop out the Igf2 gene and prevent interaction between Igf2 and enhancers. Similarly, CTCF binding and chromatin organization is necessary for activation of IFNG during T-cell differentiation. However, in the mouse β-globin locus, CTCF binding at the flanking HS5 and 3′HS1 sites results in loop formation, but the loop does not appear to be required for normal globin gene regulation in erythroid progenitor cells. These functions of CTCF all involve CTCF-mediated long-range chromatin organization, which can be manifest intra- or interchromosomally. CTCF is a very strong candidate for the role of coordinating the expression level of coding sequences with their three-dimensional position in the nucleus, apparently responding to a “code” in the DNA itself. Dynamic interactions between chromatin fibers in the context of nuclear architecture have been implicated in various aspects of genome functions. CTCF-chromatin interaction mediates gene regulation in the three-dimensional nuclear space. The “CTCF code,” explaining the mechanistic basis of how the information encrypted in DNA may be interpreted by CTCF into diverse nuclear functions.3 Furthermore, CTCF binds DNA in a methylation-sensitive manner. Therefore, DNA methylation has the potential to positively regulate gene transcription, albeit in an indirect manner, by preventing CTCF binding and thereby abolishing an enhancer block.4 In cancer, CTCF appears to function as a tumor suppressor gene.
REFERENCES
1. Phillips, J.E. & Corces, V.G.: Cell 137:1194-211, 2009
2. Burke, L.J. et al: EMBO J. 24:3291-300, 2005
3. Ohlsson, R. et al: Bioassay 32:37-50, 2010
4. Lai, A.Y. et al: J. Exp. Med. 207:1939-50, 2010
2. Burke, L.J. et al: EMBO J. 24:3291-300, 2005
3. Ohlsson, R. et al: Bioassay 32:37-50, 2010
4. Lai, A.Y. et al: J. Exp. Med. 207:1939-50, 2010
Products are for research use only. They are not intended for human, animal, or diagnostic applications.
Параметры
Cat.No.: | CP10366 |
Antigen: | Raised against recombinant human CTCF fragments expressed in E. coli. |
Isotype: | Mouse IgG1 |
Species & predicted species cross- reactivity ( ): | Human, Mouse, Rat |
Applications & Suggested starting dilutions:* | WB 1:1000 IP n/d IHC n/d ICC n/d FACS n/d |
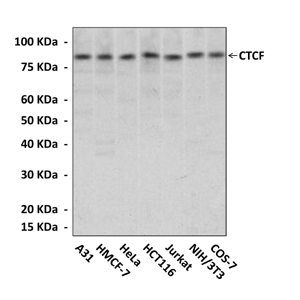
Predicted Molecular Weight of protein: | 83 kDa |
Specificity/Sensitivity: | Detects endogenous CTCF proteins in various cell lysate. |
Storage: | Store at -20°C, 4°C for frequent use. Avoid repeated freeze-thaw cycles. |
*Optimal working dilutions must be determined by end user.
Документы
Информация представлена исключительно в ознакомительных целях и ни при каких условиях не является публичной офертой