Anti-Ku80/XRCC5: Mouse Ku80/XRCC5 Antibody |
 |
BACKGROUND Ku80 (also known as XRCC5) is part of the Ku heterodimer with G22P1 (also known as Ku70), which acts with the DNA-dependent protein kinase (DNA-PK) catalytic subunit (CS) in the non-homologous end joining (NHEJ) pathway. This pathway joins the ends of DNA double-strand breaks (DSBs), whether spontaneously induced by genotoxic stress or generated by V(D)J recombination during the development of immunological diversity. The NHEJ pathway requires the sequential loading of several enzymes on both DNA ends. The first event in NHEJ-mediated repair is the association of a Ku70-Ku80 heterodimer (Ku70/80) with each DNA terminus. The Ku70/80 molecule has a ring-shaped structure, made up by the amino-terminal and central domains of both the Ku70 and the Ku80 polypeptides, which exactly fits a DNA helix in its center. The DNA-Ku complex functions as a scaffold to attract the other known NHEJ factors to the DSB. One of the enzymes that are recruited to the DNA-Ku scaffold is the DNA-dependent protein kinase catalytic subunit (DNA-PKCS). DNA-PKCS functions as a gatekeeper molecule that regulates access to the DNA termini by changing its phosphorylation status. Therefore, DNA-PKCS autophosphorylation may regulate the next steps in the NHEJ process. These next steps include the processing and joining of DNA ends. Processing enzymes prepare nonligatable DNA termini, primarily blocked ends and incompatible single-strand overhangs, for subsequent ligation by the XRCC4/ligase IV complex. The chemistry of the ligation reaction necessitates the addition of 5' phosphate groups or the removal of 3' phosphate groups by polynucleotide kinase. Processing of single-strand overhangs is performed by either filling or resection and therefore requires a polymerase or a nuclease, respectively. Several enzymes with single-strand filling capability, including polymerase lambda, polymerase mu, and terminal deoxynucleotidyltransferase, have been suggested to function as processing enzymes during NHEJ. In contrast, only one nuclease has been conclusively shown to play a role in NHEJ: the endonuclease Artemis. The NHEJ core factors DNA-PKCS, XRCC4/ligase IV, and Artemis all are attracted to a DSB by the DNA-Ku scaffold.1 NHEJ is critically important for repairing DSBs in adult mammals in which most cells are non-dividing and therefore unable to employ the alternative DNA DSB repair pathway, homologous recombination, which relies mainly on sister chromatids as templates for error-free repair. DNA DSBs are highly toxic and need to be repaired swiftly to avoid DNA degradation and cell death, or genome rearrangements resulting from erroneous end joining.2 In addition, Ku80 has also been implicated in chromosome stability and telomere maintenance, and the Ku80 gene appears to be essential for human but not rodent cell survival. Ku associates with a number of proteins involved in various processes, including replication, cell adhesion, and transcription. The diversity of Ku interactions suggests the requirements for Ku in the cell may extend beyond canonical repair.3
REFERENCES
1. Waterings, E. et al: Mol. Cell. Biol. 29:1134-42, 2009
2. Hasty, P.: J. Animal Physiol. Animal Nutrit. 93:142-3, 2009
3. Fink, L.S. et al: Cell Cycle 9:3798-3806, 2010
2. Hasty, P.: J. Animal Physiol. Animal Nutrit. 93:142-3, 2009
3. Fink, L.S. et al: Cell Cycle 9:3798-3806, 2010
Products are for research use only. They are not intended for human, animal, or diagnostic applications.
Параметры
Cat.No.: | CP10421 |
Antigen: | Raised against recombinant human Ku80 fragments expressed in E. coli. |
Isotype: | Mouse IgG1 |
Species & predicted species cross- reactivity ( ): | Human, Mouse, Rat |
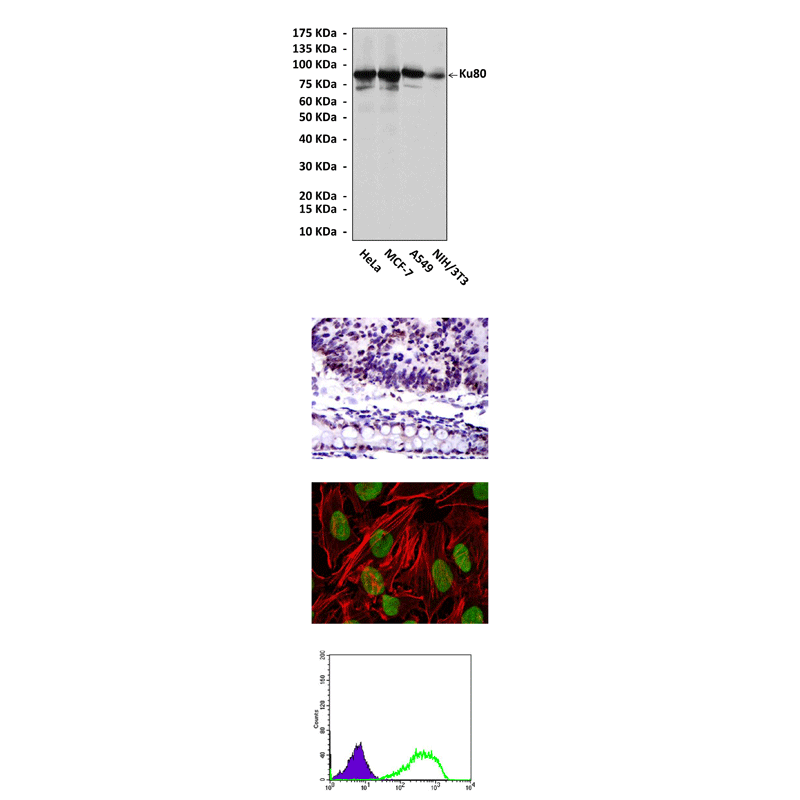
Applications & Suggested starting dilutions:* | WB 1:1000 IP 1:25 - 1:50 IHC 1:50 - 1:200 ICC 1:50 - 1:200 FACS 1:50 - 1:200 |
Predicted Molecular Weight of protein: | 86 kDa |
Specificity/Sensitivity: | Detects Ku80 proteins in various cell lysate. |
Storage: | Store at -20°C, 4°C for frequent use. Avoid repeated freeze-thaw cycles. |
*Optimal working dilutions must be determined by end user.
Документы
Информация представлена исключительно в ознакомительных целях и ни при каких условиях не является публичной офертой