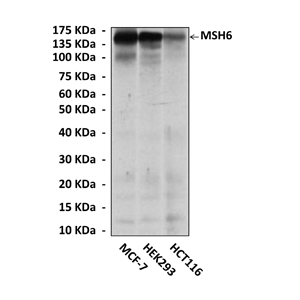
Anti-MSH6: Mouse MSH6 Antibody |
 |
BACKGROUND The mismatch repair (MMR) proteins are required to maintain genomic integrity in prokaryotes and eukaryotes, by correcting single mismatches and short unpaired regions, such as small insertions and deletions. In eukaryotes, three proteins are involved in mismatch recognition, MSH2, MSH3 and MSH6. The three proteins form two heterodimers MutSalpha (MSH2-MSH6) and MutSbeta (MSH2-MSH3). MutSalpha is thought to be involved primarily in the recognition and repair of base-base mismatches and small insertion/deletion loops. MutSbeta acts preferentially on insertion/deletion loops up to 12 nucleotides in length. MMR is tightly linked with the mismatch-binding-dependent ATPase activities of the MSH. Upon binding to the mismatch, heterodimers bend the DNA helix and shields approximately 20 base pairs. After mismatch binding, MutS alpha or beta forms a ternary complex with the MutL alpha heterodimer, which is thought to be responsible for directing the downstream MMR events, including strand discrimination, excision, and resynthesis.1 ATP binding and hydrolysis play a pivotal role in mismatch repair functions. The MSH6 contains a highly conserved helix-turn-helix domain associated with a Walker-A motif (an adenine nucleotide and magnesium binding motif) with ATPase activity.The ADP/ATP binding domain of the heterodimer and the associated ATPase activity function to regulate mismatch binding as a molecular switch. Both MSH2 and MSH6 can simultaneously bind ATP. The MSH6 subunit contains the high-affinity ATP binding site and MSH2 contains a high-affinity ADP binding site. Stable binding of ATP to MSH6 results in a decreased affinity of MSH2 for ADP, and binding to mispaired DNA stabilizes the binding of ATP to MSH6. Mispair binding encourage a dual-occupancy state with ATP bound to Msh6 and Msh2; following which there is a hydrolysis-independent sliding along DNA. Subsequent steps result in the excision of the mispaired region followed by DNA synthesis and ligation. This transition is crucial for mismatch repair. In addition, the breast cancer 1 gene (BRCA1) product is part of a large multisubunit protein complex of tumor suppressors, DNA damage sensors, and signal transducers. This complex is called BASC, for 'BRCA1-associated genome surveillance complex and MSH6 was found to be a part of this complex.2 MSH6 mutation is a commonly associated with Hereditary nonpolyposis colorectal cancer. It is also associated with some other cancers.3
Human MSH6 gene is transcriptionally upregulated 2.5 fold at late G1/early S phase while the amount of protein remains unchanged during the whole cell cycle. The promoter region has a high GC content, as well as multiple start sites. Sequence analysis of 3.9 kb of the 5'-upstream region of the MSH6 gene revealed the absence of TATAA- or CAAT-boxes. Seven consensus binding sequences for the ubiquitous transcription factor Sp1 were found in the promoter region. This factor is implicated in positioning the RNA polymerase II complex at the transcriptional start sites of promoters lacking TATA- and CAAT-boxes. The proximal promoter region of MSH6 gene also contains several consensus binding sites of the embryonic TEA domain-containing factor ETF. This transcription factor has also been reported to stimulate transcription from promoters lacking the TATA box. In addition, the trancription of MSH6 gene is downregulated by CpG methylation of the promoter region.4
Human MSH6 gene is transcriptionally upregulated 2.5 fold at late G1/early S phase while the amount of protein remains unchanged during the whole cell cycle. The promoter region has a high GC content, as well as multiple start sites. Sequence analysis of 3.9 kb of the 5'-upstream region of the MSH6 gene revealed the absence of TATAA- or CAAT-boxes. Seven consensus binding sequences for the ubiquitous transcription factor Sp1 were found in the promoter region. This factor is implicated in positioning the RNA polymerase II complex at the transcriptional start sites of promoters lacking TATA- and CAAT-boxes. The proximal promoter region of MSH6 gene also contains several consensus binding sites of the embryonic TEA domain-containing factor ETF. This transcription factor has also been reported to stimulate transcription from promoters lacking the TATA box. In addition, the trancription of MSH6 gene is downregulated by CpG methylation of the promoter region.4
REFERENCES
1. Kolodner, R.D. & Marsischky,G.T.: Curr. Opin. Genet. Dev. 9:89-96, 1999
2. Wang, Y. et al: Gene Dev. 14:927-39, 2000
3. Hunter, C. et al: Cancer Res. 66:3987-91, 2006
4. Szadkowski, M. & Jiricny, J.: Genes, chromosomes cancer. 33:36-46, 2002
2. Wang, Y. et al: Gene Dev. 14:927-39, 2000
3. Hunter, C. et al: Cancer Res. 66:3987-91, 2006
4. Szadkowski, M. & Jiricny, J.: Genes, chromosomes cancer. 33:36-46, 2002
Products are for research use only. They are not intended for human, animal, or diagnostic applications.
Параметры
Cat.No.: | CP10430 |
Antigen: | Raised against recombinant human MSH6fragments expressed in E. coli. |
Isotype: | Mouse IgG1 |
Species & predicted species cross- reactivity ( ): | Human |
Applications & Suggested starting dilutions:* | WB 1:1000 IP n/d IHC n/d ICC n/d FACS n/d |
Predicted Molecular Weight of protein: | 160 kDa |
Specificity/Sensitivity: | Detects endogenous MSH6 proteins without cross-reactivity with other family members. |
Storage: | Store at -20°C, 4°C for frequent use. Avoid repeated freeze-thaw cycles. |
*Optimal working dilutions must be determined by end user.
Документы
Информация представлена исключительно в ознакомительных целях и ни при каких условиях не является публичной офертой