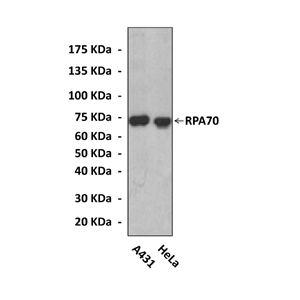
Anti-RPA70: Rabbit Replication Protein A Antibody |
 |
BACKGROUND Human Replication Protein A (hsRPA) is required for multiple cellular processes in DNA metabolism including DNA repair, replication and recombination. It binds single-stranded DNA with high affinity and interacts specifically with multiple proteins. hsRPA forms a heterotrimeric complex composed of 70-, 32- and 14-kDa subunits (henceforth RPA70, RPA32, and RPA14). RPA has a modular structure. The RPA70 subunit has four domains; the N-terminal protein–protein interaction domain (RPA70N; amino acids 1–110) and three ssDNA-binding domains (DBDs) arranged in tandem, DBD-A (amino acids 181–290), DBD-B (amino acids 301–422) and DBD-C (amino acids 436–616). DBD-C contains a conserved Cys4-type zinc-binding motif; the bound zinc modulates DNA-binding. This region has been suggested to be involved in binding to damaged DNA. The RPA32 subunit consists of three domains. The central ssDNA-binding domain, DBD-D (amino acids 43–171) is flanked at the N-terminus by a small, unstructured domain, which carries all known human RPA phosphorylation sites (RPA32N; 1–40), and at the C-terminus by a protein–protein interaction module (RPA32C; 200–270). The entire small subunit, RPA14, is folded in one structural domain. The C-terminal part of this subunit is important for trimerization. Other functions of RPA14 are currently not clear. Trimerization is mediated by three domains: DBD-C (in RPA70), DBD-D (in RPA32) and RPA14, which together form the trimerization core.1
RPA binding to ssDNA occurs via a multistep pathway and is associated with a significant conformational change. First, globular RPA binds to 8–10 nucleotides (nt) in an unstable manner. Current evidence associates this binding with DBD-A and DBD-B, the two major DNA-binding domains that harbor most of the binding activity of the full trimer. Cross-linking experiments indicate that DBD-D does not interact with ssDNA in this mode. The second step is associated with a significant conformational change to a mode with an occluded binding size of ~30 nt per trimer. The latter mode is thought to be facilitated by two minor DNA-binding domains, DBD-C and DBD-D. In this mode, DBD-A, -B, -C and -D, together directly contact between 23 and 27 nt. Several more nucleotides bridge the space between trimers bringing the occluded size up to ~30 nt. An intermediate 13–14 nt or 12–22 nt binding mode has recently been characterized. In this mode, RPA binds to ssDNA with higher affinity, as compared with the 8–10 nt mode, and does not contact ssDNA with DBD-D, as compared with the 30 nt mode.2 In addition to binding ssDNA and proteins involved in DNA metabolism RPA can destabilize double-stranded DNA (dsDNA). The destabilizing activity of RPA may play a role in the initiation of replication, as well as the denaturation of damaged DNA, in nucleotide excision repair.3
In addition, RPA interacts with various functional proteins. The N-terminal 168 residues of RPA70 form a structurally distinct domain that stimulates DNA polymerase alpha activity, interacts with several transcriptional activators including tumor suppressor p53, and during the cell cycle it signals escape from the DNA damage induced G2/M checkpoint. Moreover, RPA is phosphorylated in a cell cycle-dependent manner (during S and G2) and in response to DNA damage. Phosphorylation or mutations that add multiple negative charges to the N-terminal phosphorylation domain of RPA32 cause altered interactions with p53, T antigen, and DNA polymerase alpha/DNA primase. Thus the N-terminal phosphorylation modulates RPA activity. The phosphorylation affects DNA interactions too.4
RPA binding to ssDNA occurs via a multistep pathway and is associated with a significant conformational change. First, globular RPA binds to 8–10 nucleotides (nt) in an unstable manner. Current evidence associates this binding with DBD-A and DBD-B, the two major DNA-binding domains that harbor most of the binding activity of the full trimer. Cross-linking experiments indicate that DBD-D does not interact with ssDNA in this mode. The second step is associated with a significant conformational change to a mode with an occluded binding size of ~30 nt per trimer. The latter mode is thought to be facilitated by two minor DNA-binding domains, DBD-C and DBD-D. In this mode, DBD-A, -B, -C and -D, together directly contact between 23 and 27 nt. Several more nucleotides bridge the space between trimers bringing the occluded size up to ~30 nt. An intermediate 13–14 nt or 12–22 nt binding mode has recently been characterized. In this mode, RPA binds to ssDNA with higher affinity, as compared with the 8–10 nt mode, and does not contact ssDNA with DBD-D, as compared with the 30 nt mode.2 In addition to binding ssDNA and proteins involved in DNA metabolism RPA can destabilize double-stranded DNA (dsDNA). The destabilizing activity of RPA may play a role in the initiation of replication, as well as the denaturation of damaged DNA, in nucleotide excision repair.3
In addition, RPA interacts with various functional proteins. The N-terminal 168 residues of RPA70 form a structurally distinct domain that stimulates DNA polymerase alpha activity, interacts with several transcriptional activators including tumor suppressor p53, and during the cell cycle it signals escape from the DNA damage induced G2/M checkpoint. Moreover, RPA is phosphorylated in a cell cycle-dependent manner (during S and G2) and in response to DNA damage. Phosphorylation or mutations that add multiple negative charges to the N-terminal phosphorylation domain of RPA32 cause altered interactions with p53, T antigen, and DNA polymerase alpha/DNA primase. Thus the N-terminal phosphorylation modulates RPA activity. The phosphorylation affects DNA interactions too.4
REFERENCES
1. Bochkareva, E. et al: EMBO J. 21:1855-63, 2001
2. Lao, Y. et al: Biochem. 39:850-9, 2000
3. Treuner, K. et al: J. Mol. Biol. 259:104-12, 1996
4. Binz, S.K. et al: J. Biol. Chem 278:35584-91, 2003
2. Lao, Y. et al: Biochem. 39:850-9, 2000
3. Treuner, K. et al: J. Mol. Biol. 259:104-12, 1996
4. Binz, S.K. et al: J. Biol. Chem 278:35584-91, 2003
Products are for research use only. They are not intended for human, animal, or diagnostic applications.
Параметры
|
Cat.No.:
|
CG1526
|
|
Antigen:
|
Short peptide from human RPA70.
|
|
Isotype:
|
Rabbit IgG
|
|
Species & predicted
species cross-
reactivity ( ):
|
Human
|
|
Applications &
Suggested starting
dilutions:*
|
WB 1:10000 - 1:50000
IP 1:50
IHC n/d
ICC n/d
FACS 1:100
|
|
Predicted Molecular
Weight of protein:
|
70 kDa
|
|
Specificity/Sensitivity:
|
Detects endogenous RPA70 proteins without cross-reactivity with other family members.
|
|
Storage:
|
Store at -20°C, 4°C for frequent use. Avoid repeated freeze-thaw cycles.
|
*Optimal working dilutions must be determined by end user.
Информация представлена исключительно в ознакомительных целях и ни при каких условиях не является публичной офертой